Bringing tidy principles to a fundamental visualization for gene expressions, being on your best "behavior" for organizing your tests, and how data.table stacks up to DuckDB and polars for reshaping your data layouts.
Episode Links
Episode Links
- This week's curator: Jon Carroll - @jonocarroll@fosstodon.org (Mastodon) & @carroll_jono (X/Twitter)
- Exploring the tidyHeatmap R package
- Don't Expect That "Function Works Correctly", Do This Instead
- Comparing data.table reshape to duckdb and polars
- Entire issue available at rweekly.org/2024-W43
- tidyHeatmap: Draw heatmap simply using a tidy data frame https://stemangiola.github.io/tidyHeatmap/
- Novel App knock-in mouse model shows key features of amyloid pathology and reveals profound metabolic dysregulation of microglia https://molecularneurodegeneration.biomedcentral.com/articles/10.1186/s13024-022-00547-7
- Shiny App-Packages chapter on writing tests and specifications https://mjfrigaard.github.io/shiny-app-pkgs/test_specs.html
- WANT CLEANER UNIT TESTS? TRY ARRANGE, ACT, ASSERT COMMENTS https://jakubsob.github.io/blog/want-cleaner-test-try-arrange-act-assert/
- Super Data Science Podcast 827: Polars: Past, Present and Future, with Polars Creator Ritchie Vink https://www.superdatascience.com/podcast/827
- duckplyr: A DuckDB-backed version for dplyr https://duckplyr.tidyverse.org/
- Use the contact page at https://serve.podhome.fm/custompage/r-weekly-highlights/contact to send us your feedback
- R-Weekly Highlights on the Podcastindex.org - You can send a boost into the show directly in the Podcast Index. First, top-up with Alby, and then head over to the R-Weekly Highlights podcast entry on the index.
- A new way to think about value: https://value4value.info
- Get in touch with us on social media
- Eric Nantz: @rpodcast@podcastindex.social (Mastodon) and @theRcast (X/Twitter)
- Mike Thomas: @mike_thomas@fosstodon.org (Mastodon) and @mike_ketchbrook (X/Twitter)
- Black Feathers in the Sky - Kid Icarus: Uprising - MkVaff - https://ocremix.org/remix/OCR04200
- Cross-Examination - Phoenix Wright: Ace Attorney - PrototypeRaptor - https://ocremix.org/remix/OCR01846
[00:00:03]
Eric Nantz:
Hello, friends. We're back with a 183 of the R Weekly Highlights podcast. This is your weekly podcast. We're gonna talk about the great resources that are shared every single week on this week's Our Weekly Issue. My name is Eric Nantz, and I'm delighted you're joining us from wherever you are around the world. Already near the end of October, it's hard to believe the time is flying by. The air is crisp in the mornings as I ride my bike to my kid at school. You can feel the the chill in the air, but, nonetheless, we're heating things up here in more ways than one with this episode. I can't do that alone, of course. My, next, generator of our heat, if you will, is right here next to me virtually, Mike Thomas. Mike, how are you doing today?
[00:00:52] Mike Thomas:
I'm doing pretty well, Eric. This is the first I guess I'm spoiling it a little bit, but this is the first our weekly I've seen in a couple weeks, we where we are not discussing AI in any of the three highlights. I don't know if that means that the AI buzz has has cooled off maybe.
[00:01:03] Eric Nantz:
Or they know that we need a break from it either way. Probably both. It could be both. It could be both. But, yes, it's good to have variety in life as we say in this very show, and we got a good variety of content to talk about with you today. And our weekly, if you're new to the project, this is a terrific resource. We aggregate all the awesome use cases of R across data science and industry, academia, and new packages and updated packages and great tutorials. And we got a mix of all of the above on this one. And it has been curated this week by our good friend, Jonathan Carroll, who is also gonna be involved with helping chair for the first time ever the R pharma conference. It's not just having a, you know, US or western hemisphere based track. He is helping chair the APAC track for the Asia Pacific region. So we're very happy to, John, to have you involved with our team. He's already been hard at work preparing for that conference. But as always, with our weekly, he had tremendous help from our fellow, our weekly team members, and contributors like you all around the world of your poll request and other great suggestions.
And like I said, we're gonna heat things up initially on this very podcast, and we're gonna talk about a fundamental pillar of visualization across multiple disciplines, and that is the venerable heat map. And if you you know, as usual on a podcast, it's hard to describe everything in audio fashion. But a heat map, if you haven't seen that before, is a 2 dimensional, you might say, grid where each cell is kind of the, you know, the expression, if you will, of how large or small a quantity is. And we see this a lot in, for example, correlation analyses where you might look at all the pairwise correlations across a group of variables, and each of those combinations is a cell in the heat map with a higher correlation, which is, of course, between 01, might get either a brighter color or darker color depending on how you invert the palette and whatnot.
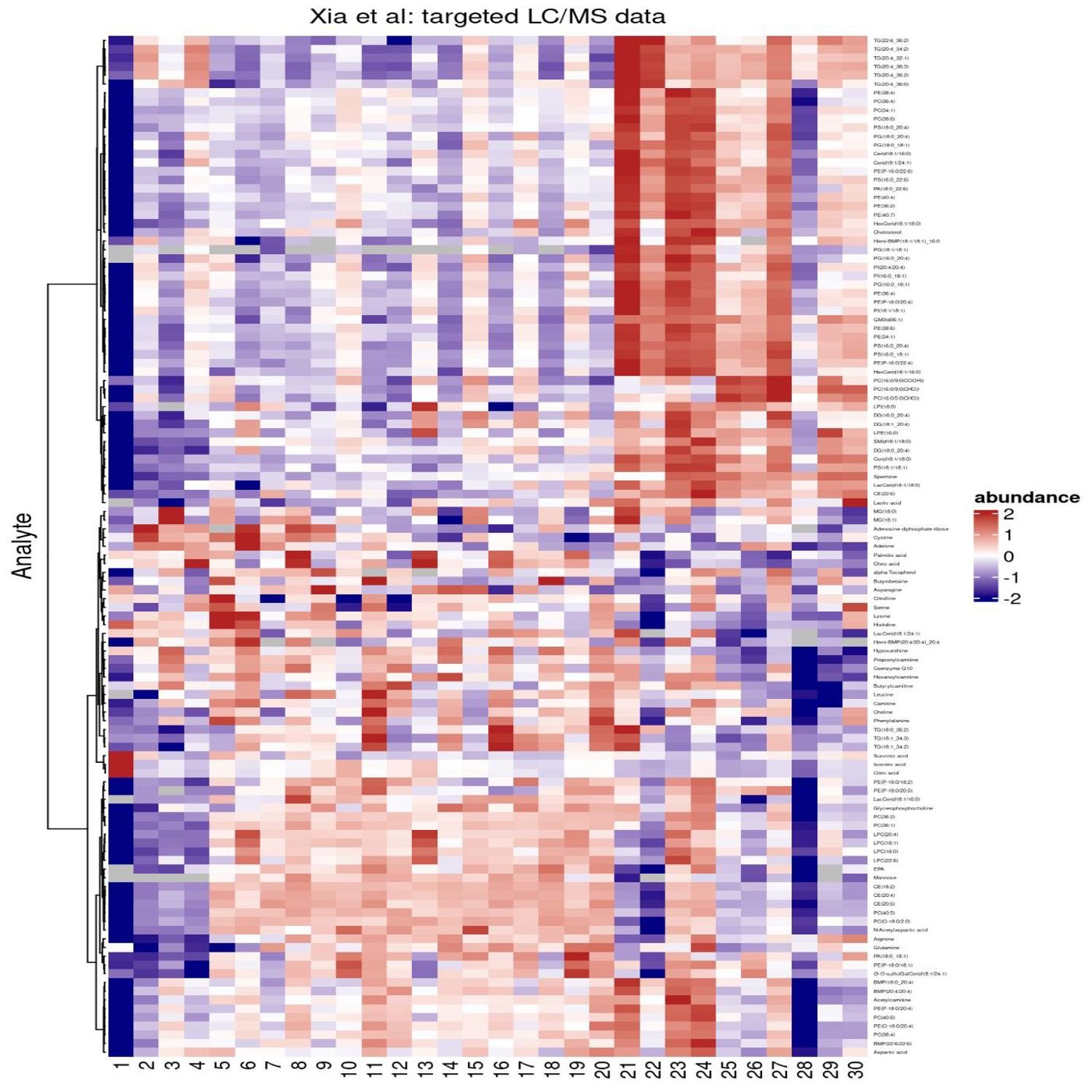
And there is another domain where heat maps are very much in play and very much a fundamental, you might say, pillar of data exploration, and that is the world of genomics and biomarker analyses. From my days many years ago dealing with the Bioconductor project, I create a lot of these heat maps often using the core set of Bioconductor packages that would have nice wrappers around the typical object types that you get in Bioconductor, which is typically the matrix. A matrix is a fundamental data input in many of the classical heat map functions. Maybe you're getting data that already isn't quite in the Bioconductor like layout, but you want to take advantage of some of these great visualization techniques like heat maps. And that's where our first highlight comes into play.
It is a blog post author by Thomas Sandman, who is a scientist at Denali Therapeutics, and I can tell who has vast experience in the world of genetic and biomarker and PKPD analyses because he is actually attempting in this blog post to recreate some very nice heat map visualizations from a recent manuscript looking at the that the effect of a specific mutation of a gene for understanding more of the pathology behind what turns out to be neurodegeneration, which many people are familiar with the Alzheimer's disease as one of the manifests of that. But there has been research in many, and I do mean many decades, on trying to find the best genes and other biomarkers to target to try and hopefully minimize the impact of this, you know, debilitating disease and hopefully even cure it altogether.
It's been a lot of research. There's been a lot of misses along the way, but there are some promising avenues in this manuscript of a few lipids and also other additional biomarkers that they did an experiment with mice to see what the gene expression would be in certain parts of their brains on this regulation of these of these genetic markers. And the heat map is, like I said, a classic way to visualize the impact, in this case, of some linear models looking at the different, you know, characteristics of these lipids after this experiment.
And so the post starts off with giving us, of course, a nice link to this manuscript, which we'll link to in the show notes as well. But this manuscript, if you do dig through it, has a little bit of a description on the methods used and some of the r packages that were used mostly from Bioconductor and, thankfully, example Excel files with the results of the stat analysis. However, there was no article shared with this, which, again, unfortunately, is kind of common in these areas. Right? You may get snippets of this for reproducibility, but you don't quite get the full picture. So with that said, the the journey begins with trying to figure out, okay, how do we get this data ready to go for a statistical analysis or a visualization, I should say, with heat maps.
The first step is to import this data from Excel, and there's some nice, I can tell this blog is written record, also got some nice call out blocks that you can expand to look at the different functions that Thomas has outlined here. Really nice use of ReadExcel and TidyR to get the data into the right shape for the eventual, heat map visualizations. Also, there was a additional spreadsheet that the researchers shared with the statistical analysis, so he's got, CSV import of that particular file as well if you just want to get the finished product of that. And to make generating heat maps easier, Thomas is spotlighting the package tidy heatmap, which is a nice front end to another package called complex heatmap, which again would expect the data going into it to be in matrix form.
But tidy heat map lets you have a more, you know, tidy ish type data where you've got your rows as observations, columns as the variables. And he's got a nice way, once you do a little filtering and a little pre processing to match kind of the groupings that went into this original manuscript's heat map visualization. It literally is a function called heat map feeding in the data frame, which variable corresponds to your rows, which variable corresponds to your column, which in this case is the sample IDs of these different mice because there are different samples taken for each for each specimen.
And then what is the value going into it? In this case, it's called an abundance measure, which is the fold change log transform of the gene expression of these markers. And right away, you got a pretty nice looking heat map along the way that looks like straight out of ggplot2. I believe it's using that under the hood, but you can do a lot more to this. And that's what the rest of the post talks about. How do we go from that very quick starting point to a heat map that more matches what the manuscript outline? So there's a few steps here involved, one of which is to change the color palette a little bit.
And there is, a set of packages that are being used here. 1 is called circlewise, which I haven't seen before, which has its color ramp 2 function to basically say, okay. For this amount of breaks in your numeric axis, use these set of colors going from navy to firebrick with the different ranges, and then you can now feed that into that original heat map call. So now you've got a heat map that definitely looks a little more like you would see in many of these manuscripts already colorized differently. And then also there is additional ways that you can put grouping annotations on top of those columns, which in this case were depicting the sample IDs.
They have an inherent grouping between those, so this is going to let you do, like, a visual split, kind of like a little bar with an annotation in the middle over each of these groups. So you can quickly see then the different types of samples based on the different type of genetic marker it was targeting. So that's already very familiar in the ggplot2 side of things with fascinating. But that's not all. You can also add custom annotations as well where there is a handy function called annotation or annotation_tile, which now under those headings that you saw above each column, you can then do like a in essence a color like legend that depicts different groups within each of these overall groups. In this case, the batch, which is these samples usually goes in a different batch for the experiment, and then also the gender of these mice. So those can be neatly colored so your eyes can quickly see then on top of this one genetic marker what were the batch the batch ID in terms of color and then also the gender associated with that. So it's a really, really handy way to visualize that grouping in a categorical fashion but with colors instead.
Lastly, but certainly not least, we got some additional processing to do, and that's where we start to look at how do you annotate additional quantitative information underneath these these additional group grouping of colors based on the cell number that these markers are coming from. So that's another handy function waiting to happen with an additional use of annotation tile and annotation point. So, again, audio won't do this much justice, but underneath those color regions, he's got little dots that that depict the cell number with a little y axis right at the left. So already done about 3 or 4 different types of variables above the columns.
And then the last part is about how the rows are organized, and this is gonna take a little more dplyr data munging magic to make sure that the groups match kind of the grouping order based on a) the expression level and then also doing a more manual grouping fashion to match kind of different overall groups that we saw in the heat map earlier in the manuscript. So all the code is reproducible. Again, this heat map function has got a lot of parameters inside. But depending on how in-depth you want to make your customizations, there's typically a parameter for it. Like I mentioned, this column grouping, the way you can organize the different rows, the way you can put these annotations together.
I never knew how much power was exposed to us with these heatmap functionality. So next time I whether I do a biomarker analysis in the future or even go to my tried and true correlation analysis, I'm gonna have to give tidy heat map a try. This looks really fun.
[00:12:53] Mike Thomas:
Yeah. This is pretty incredible. If it is ggplot under the hood, it's insane, you know, how far we can push it to customize all sorts of stuff here. I I really like sort of the approach that has been taken to to tidy up, if you will, by Thomas, the complex heat map package. If you're somebody who's more familiar with the tidyverse, I think you're gonna find the API here to be a lot more friendly. I took a look at the complex heat map, our package, and it actually has some fantastic documentation. There's, like, a whole book on using the complex heat map package. But as you mentioned earlier, Eric, sort of the the root unit that you're gonna pass to this this heat map function that starts with a capital h, we can talk about that later, as opposed to a lowercase h, is a matrix as opposed to a data frame.
And I think, you know, this complex heatmap package tries to maybe reduce the number of parameters passed to the heat map function and abstract away, some of the heavy lifting for you. But I it seems to me when it goes about doing that, that it it makes probably a lot of assumptions off the bat about how it's going to, display that data, sort of what column should be named as, things like that, how the data should be structured. And I think you just have a little bit more control with the API, at least in terms of getting started for those who are coming from a more, you know, tidyverse, background with this, new package that that Thomas has put together. So we have some clients that do some, you know, bioinformatics, biomarker type of work, and the heat map is, like, the most important tool, data visualization tool, for them to be able to use. And oftentimes, they wanna push those heat maps, as far as they can go with multiple different legends, dendograms on the side. Right? We have, also on, you know, sort of the other vertical axis of the dendogram's on the left side of the heat map, on the right side, in this example here that Thomas has. We have, for every single sort of row on the heat map, a particular label.
And those can be very, very small, difficult to see if you're not providing arguments in your API to be able to adjust all these things to try to make these heat maps, which are very complex by nature, as digestible as possible to those who are going to be making the decisions based upon the data that they're seeing in in a way that they're interpreting interpreting the heat maps that you're putting together. So I I think it's incredibly powerful that we have sort of this much control over how to develop these heat maps. I really, you know, like the the syntax that, this API has in Thomas's new package. And I think folks who are looking to take their heat maps to the next step and certainly all the folks that we work with in the the biomarker and bioinformatics space, I'm going to pass this blog post along to them because I think this is going to to save them time and be able to help them accomplish all of the little nuances that they wanna introduce into their visualizations.
[00:16:06] Eric Nantz:
And I do stand corrected, my friend. Real time correction here. I did check the packages that it depends on. It actually does not depend on ggplot2. It's it's depending on the grid system, of course, which is a fundamental foundation of ggplot2. But interestingly enough, we'll have a link to the tidy heatmap, package down site in the show notes. They also integrate with Patchwork, which is what we've talked about previously to stitch multiple grid based plots together, which now it all comes together in real time, doesn't it? Those nice, you know, annotations that we talked about, you know, going between the column labels and the actual content of the heat map itself, that seems like patchwork under the hood if I had to guess with those custom visualization types above the above the main the main area. So really, really interesting use case. I had no idea the kind of directions that that could take, but I and the yeah. Definitely check out check out the heat the tiny heat map site. There's lots of great examples on top of what Thomas has done in this blog, which is, of course, focused on the more genetic, you know, biomarker suite of things, which I'm sure many of you will be very appreciative of. But, yeah, there's lots of great examples in the vignettes that I'm looking at already, in the overview. Lots lots of things to to to digest, if you will, in your heat map exploration.
So this is going to my massive set of visualization bookmarks for future reference. Mike, we always love our continuity on our weekly hallways podcast. And last week, we were talking about a novel way to help organize maybe a many, many sets of tests that have some relationships amongst each other with a nesting approach. And if you're not familiar with that, go listen back to our previous episode 182 where we had talked about Roman Paul's announced or blog post on organizing tests with a nesting approach. There are always more than one way to do things in the r ecosystem, and our next highlight here comes us from Jacob Sobolowski, who is a r and shining developer at Appsilon, who if you haven't been following his blog, he has been all over the different ways of how he thinks about testing complex shiny applications, many thought provoking posts that he has on his blog. But in particular, this one here is talking about another approach to organize and really notes for future use, so to speak, on the different tests that you can organize that have some relationship with each other.
So to motivate this, he has a simple example, again very trivial but hopefully you can generalize it, of a simple set of expectations to test that the median function in base r is performing as expected. So in the test that call, which, again, test that is the package that's helping, you know, drive the engine behind many of the automated testing, paradigms in r that you see in package development, Shiny apps, and whatnot, It has a simple call to test underscore that, and the description reads, does median works correctly.
And so within that, there's a set of 6 expect equals in this case of the different, you know, numbers that he feeds in and does it get the actual number in the answer. Again, very very straightforward, but you are seeing that the median should, when you look at these functions, handle a few different use cases. Whereas, if it's only one element, should give you the same value back. Or if it's 2 elements, should be the average. If it's an odd number of elements, should be the middle number in sequential order. Blah blah. You you understand that. Now imagine that had been a more complex function.
Imagine that the expectations may not be as obvious at first glance when it's really trying to infer here. So his next, snippet of code is now going to write test that, but each of the tests is corresponding to those individual expectations. So like I just mentioned earlier, he's got, in this case, 4 different tests that are, you know, verifying the different behaviors based on either how many numbers are going in or, you know, in fact, that is basically it. And Revver, does the same thing for order and unordered elements.
Now that's, again, a perfectly valid approach. It is illustrating that test that when you put these descriptions inside, that test that function is named that for a reason. You kind of read it as test that and then something. However, in this case, it's like the median should return the same value if the vector only has one element. You you get the idea there. But here comes the kicker. In TestDaT, there are additional functions to organize your tests even further. And this is getting into a paradigm that admittedly is not quite comfortable to me just yet called behavior driven development or BDD syntax and test that for I'm not sure how many releases up to this point has spurred the use of a function called a set of functions called describe and it.
So in this last snippet of code, Jacob has an overall describe call that just simply says median. And then within it are a series of it calls and then the description in these it function calls is kind of the behavior itself that he's trying to test, where we have about the same value if it's only one element, the average of 2 values if it has an even number, the middle value in sequential order if it's an odd number of elements, and that it's the same value for ordered and unordered elements. I've seen this once before when I in Pasa Comps in recent years, I've had some interesting conversation with Martin Frigard, who has written a book called Shiny app as packages or something to that effect. But he has a specific chapter on behavior driven development in in light of specifications and requirements and how your test that test can kinda mirror that approach.
Admittedly, I have not had enough practice of describing it, no pun intended, in this workflow, but maybe it is quite helpful in your use case to have, again, another approach to how you organize related expectations into an overall testing block. So it is kind of a paradigm shift. Right? We could have either the nesting approach that we saw last week with the test that and multiple test that calls inside or the describe, and then these it functions are gonna have the expectations directly. Which is right for you? I don't know. Mike, what's your take on this?
[00:23:34] Mike Thomas:
This is the 2nd week in a row, I think, maybe 2 out of 3 weeks where I have found things in the test that package that I had no idea. Literally no idea existed. And, Eric, as I am, I know you are, we're very organized people and probably fairly opinionated about how we organize our code as well. I am open to this. I think if you were looking for a hot take on why we shouldn't consider this and and why the old way is the best, I'm not sure you're gonna get it from me right now. And it might be still because I'm I'm reeling from, learning about this new describe it paradigm.
But just like I was sort of blown away by the organization in the in the blog post around, nesting your tests last week or the week before. I'm similarly kinda blown away by this additional functionality here where we can leverage this describe it paradigm. I'm actually even more sort of not necessarily blown away, but but more keen on the the final sentence of this blog post. It says, if you wanna push test readability even further, check out how we can use the arrange, act, assert pattern to achieve that. And if you take a look at that link it's another nice short blog post as well by by Jacob. And, I I really like sort of the concept around how he he specifies arranging, the inputs sort of acting and and evaluating those inputs, and then passing those evaluated values to a set of test assertions after the fact. And, again, in that blog post, he's usually I know this this one isn't part of the the rweekly, but it's it's a cousin. It's close enough.
Again, he's leveraging that describe it paradigm. I think it's sort of I don't know. The more I look at it, it looks like it's it's sort of, you know, 6 in 1, half dozen in the other. I'm not sure how much of a difference it it makes. I I think it's just a matter of of preference, you know, if somebody on my team decided that they wanted to use describe it versus, you know, the traditional test that, verb from the test that package. I don't think I would I would care too much either way because I think it's it's legible both ways. I think it's fairly similar, in approach. It's it's sort of just different verbiage, if you will.
But it's very interesting for me, you know, to to know that this exists, and see that potentially maybe there there might be a use case for me in the future to try to adopt them. I'm just gonna stick one toe in the water, I think, for now. You know, I was taking a look at the some of the most recent packages that Hadley Wickham has put together, Elmer being one of them. And I wanted to take a look at the unit tests in there to see if Hadley was leveraging, you know, the older type of framework that traditional test that framework or or leveraging this describe and it paradigm.
And it looks like Hadley is still utilizing the old ways, if you will. So not necessarily adopted the describe it functionality, but I believe he has or had at one point in time a large hand in, authoring and making these decisions around the the test that package. Don't quote me on that. So I imagine perhaps, he was involved in maybe a pull request or 2 that that included this describe it functionality. So I was interested to see, you know, if the if he was going to be one of the folks who had also adopted sort of this new paradigm for code organization purposes purposes because I know that Hadley preaches, you know, good software development practices and and trying to articulate your code as as best as possible.
So I guess at the end of the day here, interested to see that we have these two new functions that I did not know about in the test that package. Am I all in on it? No. But I'm not all out on it either.
[00:27:38] Eric Nantz:
And I've I'm giving this the old college try as they say with this, app at work that I've just put in the production now. I took this behavior driven approach, describe its syntax for the the test. I will admit it it felt more cumbersome than right initially, but I'm thinking the payoff is not so much for me personally in the short term. The payoff hopefully is if I get additional developers to help me with this app as a, quote, unquote, scales, buzzword for all you industry folks out there, where I might need some help to orient somebody relatively quickly over. It's a dedicated member of my team or if it's contracting resource and whatnot.
And so being able to read that more verbose, but yet under you know, might say legible, might say more digestible way of organizing the test, then, hopefully, it makes it easier for them to write tests in that similar framework. And then we all can kinda have a common, you might say, style that we can gravitate towards for that particular project. Not too dissimilar to the whole paradigm of a given project having the same code style for all of its, you know, coding files, which, again, I can speak from experience when teams don't adhere to that. There'd be dragons when you do code reviews, and I won't say that for another time. That's my hot tech.
But I I think this is I think for future proofing things, I could see this being quite valuable, especially in the other context that I was dealing with. Right? I tried to I tried to have an attempt back to Martin's book that I'm gonna put in the show notes here of really articulating those user stories if you wanna use the agile methodology into what I'm actually trying to accomplish in that test. So if there's a, quote, unquote, project manager that wants to see how I how I assess all those different user stories even though the basically, that project manager is me for this particular project. But let's say, hypothetically, it was a more larger scale project. It was a good practice to see how it goes. So, again, it didn't feel comfortable yet, but maybe the proof is a year from now. So ask me a year from now, Mike. We'll see if I take changes.
[00:29:52] Mike Thomas:
Put it on my calendar. Oh, I don't think so.
[00:30:09] Eric Nantz:
And over the course of the last, you know, might say year or or so, we have seen an influx of really top notch resources and discussions in the in the r community as a whole with respect to the data dot table package. It's been around for many years. But thanks to recent NSF grant, they have had some real robust efforts to get the word out and also putting in the different situations and different context so that users that are new to the package can really understand what are the benefits of data dot table in their daily workflows. And so our last highlight is coming from the our data table community blog, which we've talked about in previous highlights.
In this case, the post is coming from Toby Dylan Hocking, who is a statistical researcher and the director of the LASSO lab at the University of Sherbrooke, which I believe is in Canada. And he has a very comprehensive post here about the comparison of data dot table functions for reshaping your data as compared to 2 other additional packages in your ecosystem. We're gaining a lot of momentum lately, which are DuckDV and Polars. So first, let's set the stage a little bit here because especially if you're very familiar with with database operations or if you're not so much like I was before, say, a few years ago, there is somewhat different terminology between what we use and, like, the tidyverse type of explanations of reshaping or in general data manipulation and the database equivalence of this. So when we talk about a long format, we are talking about not many columns but many rows in your dataset.
And often, there are groups of these rows based on, say, a variable type or a feature or whatnot. In SQL lingo, that's called unpivoted. That was new to me a while ago. Versus the wide format when you have one record, but then the columns are representing maybe different variables, and they are literally specific to that variable. You have, again, many columns, potentially fewer rows. That is called pivoted in the SQL lingo. Again, new to me, but that's where we're we're operating on where a lot of times, you would see in the community blog posts about benchmarks talking about the classical SQL like operations, like filtering, adding new variables, group summaries, or whatnot. But now we're literally turning it on its head, so to speak, by looking at the different ways we can change our data layout.
So the first part of the post is talking about just how data dot table accomplishes this, and this terminology is actually familiar from, I would say, previous language and dplyr for some of these other operations. But first, we will talk about going from a wide format to a long format and that unpivoting operation, if you will. And in data dot table, there's a function called melt for that, which if you're familiar with tidr back in the day, there was functions called melt and cast, I believe, which has now been changed to pivot longer and pivot wider, but that may be familiar for the tidr veterans out there. What was interesting about data dot table's take on this on this melt function is, in this example based on the iris dataset, when you deter when you want to say what are your variable that are variable or variables determining kind of the grouping of these observations, you can actually have multiple columns generated at once.
So in this example here, going from the the iris dataset, which has in wide format columns like sepal. Width, length sepal. Width, petal. Length, petal. Width, this melt function is taking a measure. Vars argument where you can feed in this measure function the names of 1 or more variables that are defined in these groupings and either a separator in the original name of the column or, down later in the example, regex to get to those column names. That, I must say, is quite handy and eliminates a post processing step, which, as Toby talks about later on, you need additional post processing to accomplish that that same step in both polars and DuckDV.
So there's already kind of a nice concise syntax, you might say, advantage at least at first glance with data dot table going from wide to long. And it's got some example visuals of what you might do with that data. But next, how do we accomplish this in Polars? And if you haven't heard of Polars before, this is a new binding to Rust for very efficient database data like operation and tidying operations. And yes, they do support reshaping or re pivoting these data sets as well. And that is a function called unpivot going back to that SQL language. But in the example snippet, you will see that at that the first attempt you cannot do more than one variable for that grouping of the observation.
So you would have to do that in post processing afterwards, which is not difficult. But, again, it's just an extra step. But, of course, it can be done. Lastly, for DuckDV via SQL queries, you can use the unpivot SQL keyword and then feeding into what are the variables that are going in, what are what's the name of the variable that's going to define that grouping of the long observations, what's the value the column name that you wanna use as, like, the numeric result. Once again, in this case, DuckDb cannot do that that nice convenience of 2 variables at once.
You have to do that in post processing as well. So there's example snippets and and Toby's code of the blog post here to talk about that additional post processing. That that is, again, all achievable. There is another example where you can reshape into multiple columns, and that might be helpful for different types of visualizations you want to conduct. And in this case, doing an additional column for or additional columns both for sepal and petal, but then there's a grouping variable called dim, which determines if it's the length or width. So data dot table has another way of doing that as well.
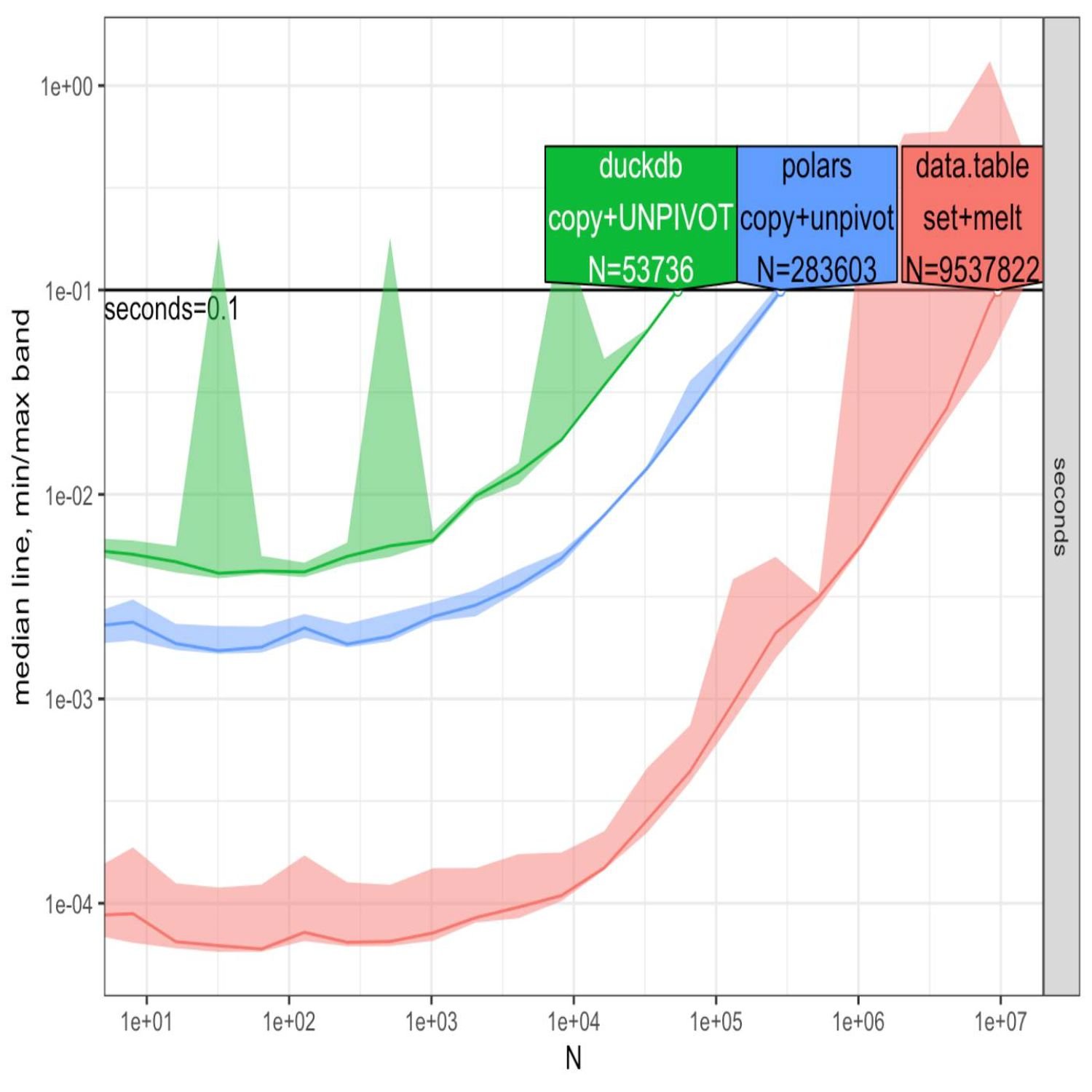
Again, the measure function comes into play there. You can see the example in the in the blog post. And then comes the comparisons for this operation. This is where my eyes open a little bit. So Toby does, a call to a an interesting func a package called a time. I'm not as familiar with this as I am with, like, the benchmark or a bench package, but he is taking a set of n values representing the number of rows in this, like, fictitious high dimensional Iris data set doing just some resampling and runs 3 different functions: 1 with the DuckDb on pivot, 2nd with the polars on unpivot, and lastly with data dot table and melt.
And across these cases, the ggplot that's put in the blog post clearly illustrates that the data dot table approach is definitely faster in this initial benchmark here than either of the other ones, especially as the end values get large. In particular, this was surprising to me, DuckDb had the worst performance by a pretty healthy margin when the end values got to, like, a 1000 rows and above. Now, again, maybe this isn't so surprising if DuckDb, of course, is based on the column type representation of data. Maybe it just isn't as optimized for these transposing operations. That very well could be a play. I'm still learning the ropes on the internals of it.
But that was interesting on top of the convenience function that data dot table has with MELT to get those multiple variables at once. It is showing, at least in this benchmark, a speedier process, especially as the number of rows increase. Now, again, maybe practically speaking, that won't be a huge impact to your workflows, but it was thought provoking nonetheless. But he does acknowledge that there's a lot of confounding in these comparisons, so it may not be quite apples to apples because of the different post processing that comes into play. And then when that is taking place, there are some additional visualizations when he kind of teases that out a bit that show the differences in even more, more fashion than what you saw in the previous plot. So interesting thought provoking exercise, but that's not the only operation, Mike, because we can also go the other way around. We can go from long to wide. So why don't you take us through that journey here? Exactly. If you are,
[00:39:50] Mike Thomas:
for some strange nonanalytics reason, looking to take your nice long data and reshape it into to wider data. And I shouldn't say that. There are some R packages that maybe it makes more sense to have each, you know, sort of sort of, a wider dataset, you know, representing your categorical data in multiple columns instead of a single column. But for me, this is a use case, that I face less often than going from from wide to long. But for those particular use cases where you do need to go long to wide, the function that you're gonna be using from data dot table is called dcast, and they provide some great examples here, Toby does, of how exactly to do that. Again, you can leverage a separator, if you would like to do that.
In this case, they're using that that period separator again, and the code is is fairly similar, to what you saw in the Melt code and allows you to pivot that that long column into a wider data frame, with multiple columns in it. If you are using polars, the method that you're going to be using there is literally called pivot. And, again, you know, the syntax is is pretty similar. You're sort of doing the opposite of what you did, when you were going from wide to long format. And then lastly, if you are using DuckDb, you're going to again use, the same named function as in Polaris. It's it's called pivot, the pivot command, which can be used to, as they say, recover the original iris data, the way that it is in wider format as opposed to long format.
And the SQL there is is pretty straightforward to look at especially compared to what we just walked through on the unpivot side of the equation. So drum roll, what everybody cares about here, right, is the benchmark comparison analysis. And this one goes a little bit inverse of the, wide to long approach such that, the engine, if you will, that has the best performance appears to be DuckDb, then closely followed by polars and then followed thereafter by data dot table. These benchmark lines that they have here are all pretty tight. I I wanna mention that that there's probably, you know, a much tighter a much tighter gaps between the 3 benchmarks here, across the 3 different engines, compared to what we saw in the wide to long approach.
So your experience is probably gonna be minimal, up until you start really scaling up to to datasets larger than, it looks like, you know, 24,000,000 rows roughly is is what DuckDV was able to handle in 0.1, seconds or or 10 milliseconds, if you will. So that was, you know, interesting to see sort of the benchmark there flip. One of the things that, Eric, I wanted to talk about here that I was curious about is I know and this is from listening to, especially, a recent podcast, not in the super data science podcast, with the author of the Polars Library, and I know DuckDV has some of this too, is both Polars and I think DuckDV have an option to do sort of a what's called a query plan and maybe lazy execution of, you know, chained functions that you might have together. Right? So we were talking about before where you can't do, you know, both in an unpivot and, you know, use a a separator in a value to split out additional columns all in a single function. You would have to chain that together in a couple different functions, in polars.
And I believe, you know, the way that polars works is depending on the default, and don't quote me on this, you know, it may default to what's called, like, eager evaluation, which is, you know, actually running those functions sort of in the order that they are written. But it provides you the option through some sort of binary flag to evaluate, your your query, if you will, or the code that you've written lazily, which means that behind the scenes, polars will put together a an optimal query plan that it believes will execute your code in the the most efficient and and fastest, if you will, way possible. And I imagine that DuckDb does some of this too.
And I also imagine that that query plan may take some time, right, under the hood to execute. So if we're only looking at these benchmarks up to 0.1 seconds, you know, I'd be interested in taking a look at datasets that are are maybe even larger, right, in the order of, like, a 100000000 rows, something like that or or larger. And seeing, you know, how these benchmarks compare after that at 0.1 second threshold to see if things change after the query planning, you know, algorithm is essentially run and the the time that it takes to do that has finished. And then we're really, you know, sort of just running, the actual code to or the actual query to do some of this data reshaping. So I'd be interested to see these benchmarks, I guess, you know, a little bit further out, extrapolate it a little bit further out in both time and magnitude of the dataset.
But, just an interesting thought that I had upon reading this blog post and it's it's super interesting to me, you know, where these, different engines sort of beat one another, if you will. You know, at the end of the day, if if you can't wait the extra 0.1 seconds or or or whatever it is, you know, half a second, just because you wanna use your your favorite engine. I guess it depends on your use case if you're standing up an API or something like that and you need response times as quick as possible and the data needs to be pivoted or or unpivoted, this is a great blog post to check out. But in sort of a general analytics, you know, approach here to to your day to day analysis.
The really cool thing I think about the ecosystem right now is is benchmarks are are getting to be fairly negligible, across most normal sized datasets.
[00:46:04] Eric Nantz:
That's a very, very fair assessment. And also with the advent of our computing technology with the processes processor cores going up and everything like that, we are seeing that for a data science workflow where let let's be real here. As much as the advancements in cloud happen, we know a lot of data science happens on people's laptops as well that with these compiled, you know, you know, I say back ends, wherever going with data dot table really based in c, and then you got polars based in Rust, the number, you know, getting a lot of attention in terms of performance. And, of course, DuckDb with its bindings. You are seeing this experience of these type of datasets. You you've got great choices here. And on that topic, like, query planning, if you want a way in especially in the DuckDV landscape, kind of get to what that query language actually looks like, I'll put a plug in the show notes for the Duckplier package, which had a CRAN release a few months ago because they have a handy function called explain that literally shows in kind of a plain text printout all the different operations that are about to be conducted. But it's doing it in an optimized order, I believe, to get the best performance as DuckDBC's fit for that particular operation, which then you can use with your familiar dplyr verbs of, like, mutate and summarize and whatnot. Again, those kind of classical operations that we've seen, you know, for for those type of work. So that was an interesting learning for me.
I do think the concept of the different data layouts is a frontier that I haven't seen as much attention on. So I'm certainly appreciative of Toby's post here to highlight, you know, the different ways that this can be accomplished. And I think, again, your use case may determine different things. I want, you know, for my back ends to have very minimal dependencies. And with that, to be able to fold into potentially a Shiny app for very quick operations about hogging the user's memory, for that particular app session. Those are my biggest, might say, criteria, if you will, as I'm looking at these.
And so I may have a use case where DuckDV does exactly what I need. There may be another use case for data dot table is king based on existing workflows. So, again, choice is gray here. Like you, Mike, I would love to see this expand in some of the different benchmarks and different scenarios to see where that plays out. But you can experiment with all these and kinda see what best fits for you. But, again, really comprehensive post by Toby here. And, again, it really shows that this funding that they're getting from this NSF grant is being put to great use to spread the word out about these different the different ways that we can accomplish these very common tasks in data science.
Well, we're we're running a bit well on time here on this episode. So we're gonna wrap things up after that great discussion of the highlights. But if you wanna get in touch with us further, we have multiple ways of doing that. You can send us a quick note in a contact page in our episode show notes. You can also send us a fun little boost along the way if you have a modern podcast app, or you can get in touch with us on social media. I am mostly on Mastodon these days, Yvette R podcast at podcastindex.social. Also find me on LinkedIn. Search my name and find me there. And, Mike, working on wisdom has got a hold of you.
[00:49:35] Mike Thomas:
Yep. You can find me on Mastodon at mike_thomas@fostodon.org, or you can find me on LinkedIn by searching Catchbrook Analytics, k e t c h b r o o k, to see what I'm up to lately.
[00:49:48] Eric Nantz:
Awesome stuff, my friend. Thanks again for a great episode, and thanks again for all of you tuning in from wherever you are. Have a great day, everybody, and we'll hopefully see you back here next week.
Hello, friends. We're back with a 183 of the R Weekly Highlights podcast. This is your weekly podcast. We're gonna talk about the great resources that are shared every single week on this week's Our Weekly Issue. My name is Eric Nantz, and I'm delighted you're joining us from wherever you are around the world. Already near the end of October, it's hard to believe the time is flying by. The air is crisp in the mornings as I ride my bike to my kid at school. You can feel the the chill in the air, but, nonetheless, we're heating things up here in more ways than one with this episode. I can't do that alone, of course. My, next, generator of our heat, if you will, is right here next to me virtually, Mike Thomas. Mike, how are you doing today?
[00:00:52] Mike Thomas:
I'm doing pretty well, Eric. This is the first I guess I'm spoiling it a little bit, but this is the first our weekly I've seen in a couple weeks, we where we are not discussing AI in any of the three highlights. I don't know if that means that the AI buzz has has cooled off maybe.
[00:01:03] Eric Nantz:
Or they know that we need a break from it either way. Probably both. It could be both. It could be both. But, yes, it's good to have variety in life as we say in this very show, and we got a good variety of content to talk about with you today. And our weekly, if you're new to the project, this is a terrific resource. We aggregate all the awesome use cases of R across data science and industry, academia, and new packages and updated packages and great tutorials. And we got a mix of all of the above on this one. And it has been curated this week by our good friend, Jonathan Carroll, who is also gonna be involved with helping chair for the first time ever the R pharma conference. It's not just having a, you know, US or western hemisphere based track. He is helping chair the APAC track for the Asia Pacific region. So we're very happy to, John, to have you involved with our team. He's already been hard at work preparing for that conference. But as always, with our weekly, he had tremendous help from our fellow, our weekly team members, and contributors like you all around the world of your poll request and other great suggestions.
And like I said, we're gonna heat things up initially on this very podcast, and we're gonna talk about a fundamental pillar of visualization across multiple disciplines, and that is the venerable heat map. And if you you know, as usual on a podcast, it's hard to describe everything in audio fashion. But a heat map, if you haven't seen that before, is a 2 dimensional, you might say, grid where each cell is kind of the, you know, the expression, if you will, of how large or small a quantity is. And we see this a lot in, for example, correlation analyses where you might look at all the pairwise correlations across a group of variables, and each of those combinations is a cell in the heat map with a higher correlation, which is, of course, between 01, might get either a brighter color or darker color depending on how you invert the palette and whatnot.
And there is another domain where heat maps are very much in play and very much a fundamental, you might say, pillar of data exploration, and that is the world of genomics and biomarker analyses. From my days many years ago dealing with the Bioconductor project, I create a lot of these heat maps often using the core set of Bioconductor packages that would have nice wrappers around the typical object types that you get in Bioconductor, which is typically the matrix. A matrix is a fundamental data input in many of the classical heat map functions. Maybe you're getting data that already isn't quite in the Bioconductor like layout, but you want to take advantage of some of these great visualization techniques like heat maps. And that's where our first highlight comes into play.
It is a blog post author by Thomas Sandman, who is a scientist at Denali Therapeutics, and I can tell who has vast experience in the world of genetic and biomarker and PKPD analyses because he is actually attempting in this blog post to recreate some very nice heat map visualizations from a recent manuscript looking at the that the effect of a specific mutation of a gene for understanding more of the pathology behind what turns out to be neurodegeneration, which many people are familiar with the Alzheimer's disease as one of the manifests of that. But there has been research in many, and I do mean many decades, on trying to find the best genes and other biomarkers to target to try and hopefully minimize the impact of this, you know, debilitating disease and hopefully even cure it altogether.
It's been a lot of research. There's been a lot of misses along the way, but there are some promising avenues in this manuscript of a few lipids and also other additional biomarkers that they did an experiment with mice to see what the gene expression would be in certain parts of their brains on this regulation of these of these genetic markers. And the heat map is, like I said, a classic way to visualize the impact, in this case, of some linear models looking at the different, you know, characteristics of these lipids after this experiment.
And so the post starts off with giving us, of course, a nice link to this manuscript, which we'll link to in the show notes as well. But this manuscript, if you do dig through it, has a little bit of a description on the methods used and some of the r packages that were used mostly from Bioconductor and, thankfully, example Excel files with the results of the stat analysis. However, there was no article shared with this, which, again, unfortunately, is kind of common in these areas. Right? You may get snippets of this for reproducibility, but you don't quite get the full picture. So with that said, the the journey begins with trying to figure out, okay, how do we get this data ready to go for a statistical analysis or a visualization, I should say, with heat maps.
The first step is to import this data from Excel, and there's some nice, I can tell this blog is written record, also got some nice call out blocks that you can expand to look at the different functions that Thomas has outlined here. Really nice use of ReadExcel and TidyR to get the data into the right shape for the eventual, heat map visualizations. Also, there was a additional spreadsheet that the researchers shared with the statistical analysis, so he's got, CSV import of that particular file as well if you just want to get the finished product of that. And to make generating heat maps easier, Thomas is spotlighting the package tidy heatmap, which is a nice front end to another package called complex heatmap, which again would expect the data going into it to be in matrix form.
But tidy heat map lets you have a more, you know, tidy ish type data where you've got your rows as observations, columns as the variables. And he's got a nice way, once you do a little filtering and a little pre processing to match kind of the groupings that went into this original manuscript's heat map visualization. It literally is a function called heat map feeding in the data frame, which variable corresponds to your rows, which variable corresponds to your column, which in this case is the sample IDs of these different mice because there are different samples taken for each for each specimen.
And then what is the value going into it? In this case, it's called an abundance measure, which is the fold change log transform of the gene expression of these markers. And right away, you got a pretty nice looking heat map along the way that looks like straight out of ggplot2. I believe it's using that under the hood, but you can do a lot more to this. And that's what the rest of the post talks about. How do we go from that very quick starting point to a heat map that more matches what the manuscript outline? So there's a few steps here involved, one of which is to change the color palette a little bit.
And there is, a set of packages that are being used here. 1 is called circlewise, which I haven't seen before, which has its color ramp 2 function to basically say, okay. For this amount of breaks in your numeric axis, use these set of colors going from navy to firebrick with the different ranges, and then you can now feed that into that original heat map call. So now you've got a heat map that definitely looks a little more like you would see in many of these manuscripts already colorized differently. And then also there is additional ways that you can put grouping annotations on top of those columns, which in this case were depicting the sample IDs.
They have an inherent grouping between those, so this is going to let you do, like, a visual split, kind of like a little bar with an annotation in the middle over each of these groups. So you can quickly see then the different types of samples based on the different type of genetic marker it was targeting. So that's already very familiar in the ggplot2 side of things with fascinating. But that's not all. You can also add custom annotations as well where there is a handy function called annotation or annotation_tile, which now under those headings that you saw above each column, you can then do like a in essence a color like legend that depicts different groups within each of these overall groups. In this case, the batch, which is these samples usually goes in a different batch for the experiment, and then also the gender of these mice. So those can be neatly colored so your eyes can quickly see then on top of this one genetic marker what were the batch the batch ID in terms of color and then also the gender associated with that. So it's a really, really handy way to visualize that grouping in a categorical fashion but with colors instead.
Lastly, but certainly not least, we got some additional processing to do, and that's where we start to look at how do you annotate additional quantitative information underneath these these additional group grouping of colors based on the cell number that these markers are coming from. So that's another handy function waiting to happen with an additional use of annotation tile and annotation point. So, again, audio won't do this much justice, but underneath those color regions, he's got little dots that that depict the cell number with a little y axis right at the left. So already done about 3 or 4 different types of variables above the columns.
And then the last part is about how the rows are organized, and this is gonna take a little more dplyr data munging magic to make sure that the groups match kind of the grouping order based on a) the expression level and then also doing a more manual grouping fashion to match kind of different overall groups that we saw in the heat map earlier in the manuscript. So all the code is reproducible. Again, this heat map function has got a lot of parameters inside. But depending on how in-depth you want to make your customizations, there's typically a parameter for it. Like I mentioned, this column grouping, the way you can organize the different rows, the way you can put these annotations together.
I never knew how much power was exposed to us with these heatmap functionality. So next time I whether I do a biomarker analysis in the future or even go to my tried and true correlation analysis, I'm gonna have to give tidy heat map a try. This looks really fun.
[00:12:53] Mike Thomas:
Yeah. This is pretty incredible. If it is ggplot under the hood, it's insane, you know, how far we can push it to customize all sorts of stuff here. I I really like sort of the approach that has been taken to to tidy up, if you will, by Thomas, the complex heat map package. If you're somebody who's more familiar with the tidyverse, I think you're gonna find the API here to be a lot more friendly. I took a look at the complex heat map, our package, and it actually has some fantastic documentation. There's, like, a whole book on using the complex heat map package. But as you mentioned earlier, Eric, sort of the the root unit that you're gonna pass to this this heat map function that starts with a capital h, we can talk about that later, as opposed to a lowercase h, is a matrix as opposed to a data frame.
And I think, you know, this complex heatmap package tries to maybe reduce the number of parameters passed to the heat map function and abstract away, some of the heavy lifting for you. But I it seems to me when it goes about doing that, that it it makes probably a lot of assumptions off the bat about how it's going to, display that data, sort of what column should be named as, things like that, how the data should be structured. And I think you just have a little bit more control with the API, at least in terms of getting started for those who are coming from a more, you know, tidyverse, background with this, new package that that Thomas has put together. So we have some clients that do some, you know, bioinformatics, biomarker type of work, and the heat map is, like, the most important tool, data visualization tool, for them to be able to use. And oftentimes, they wanna push those heat maps, as far as they can go with multiple different legends, dendograms on the side. Right? We have, also on, you know, sort of the other vertical axis of the dendogram's on the left side of the heat map, on the right side, in this example here that Thomas has. We have, for every single sort of row on the heat map, a particular label.
And those can be very, very small, difficult to see if you're not providing arguments in your API to be able to adjust all these things to try to make these heat maps, which are very complex by nature, as digestible as possible to those who are going to be making the decisions based upon the data that they're seeing in in a way that they're interpreting interpreting the heat maps that you're putting together. So I I think it's incredibly powerful that we have sort of this much control over how to develop these heat maps. I really, you know, like the the syntax that, this API has in Thomas's new package. And I think folks who are looking to take their heat maps to the next step and certainly all the folks that we work with in the the biomarker and bioinformatics space, I'm going to pass this blog post along to them because I think this is going to to save them time and be able to help them accomplish all of the little nuances that they wanna introduce into their visualizations.
[00:16:06] Eric Nantz:
And I do stand corrected, my friend. Real time correction here. I did check the packages that it depends on. It actually does not depend on ggplot2. It's it's depending on the grid system, of course, which is a fundamental foundation of ggplot2. But interestingly enough, we'll have a link to the tidy heatmap, package down site in the show notes. They also integrate with Patchwork, which is what we've talked about previously to stitch multiple grid based plots together, which now it all comes together in real time, doesn't it? Those nice, you know, annotations that we talked about, you know, going between the column labels and the actual content of the heat map itself, that seems like patchwork under the hood if I had to guess with those custom visualization types above the above the main the main area. So really, really interesting use case. I had no idea the kind of directions that that could take, but I and the yeah. Definitely check out check out the heat the tiny heat map site. There's lots of great examples on top of what Thomas has done in this blog, which is, of course, focused on the more genetic, you know, biomarker suite of things, which I'm sure many of you will be very appreciative of. But, yeah, there's lots of great examples in the vignettes that I'm looking at already, in the overview. Lots lots of things to to to digest, if you will, in your heat map exploration.
So this is going to my massive set of visualization bookmarks for future reference. Mike, we always love our continuity on our weekly hallways podcast. And last week, we were talking about a novel way to help organize maybe a many, many sets of tests that have some relationships amongst each other with a nesting approach. And if you're not familiar with that, go listen back to our previous episode 182 where we had talked about Roman Paul's announced or blog post on organizing tests with a nesting approach. There are always more than one way to do things in the r ecosystem, and our next highlight here comes us from Jacob Sobolowski, who is a r and shining developer at Appsilon, who if you haven't been following his blog, he has been all over the different ways of how he thinks about testing complex shiny applications, many thought provoking posts that he has on his blog. But in particular, this one here is talking about another approach to organize and really notes for future use, so to speak, on the different tests that you can organize that have some relationship with each other.
So to motivate this, he has a simple example, again very trivial but hopefully you can generalize it, of a simple set of expectations to test that the median function in base r is performing as expected. So in the test that call, which, again, test that is the package that's helping, you know, drive the engine behind many of the automated testing, paradigms in r that you see in package development, Shiny apps, and whatnot, It has a simple call to test underscore that, and the description reads, does median works correctly.
And so within that, there's a set of 6 expect equals in this case of the different, you know, numbers that he feeds in and does it get the actual number in the answer. Again, very very straightforward, but you are seeing that the median should, when you look at these functions, handle a few different use cases. Whereas, if it's only one element, should give you the same value back. Or if it's 2 elements, should be the average. If it's an odd number of elements, should be the middle number in sequential order. Blah blah. You you understand that. Now imagine that had been a more complex function.
Imagine that the expectations may not be as obvious at first glance when it's really trying to infer here. So his next, snippet of code is now going to write test that, but each of the tests is corresponding to those individual expectations. So like I just mentioned earlier, he's got, in this case, 4 different tests that are, you know, verifying the different behaviors based on either how many numbers are going in or, you know, in fact, that is basically it. And Revver, does the same thing for order and unordered elements.
Now that's, again, a perfectly valid approach. It is illustrating that test that when you put these descriptions inside, that test that function is named that for a reason. You kind of read it as test that and then something. However, in this case, it's like the median should return the same value if the vector only has one element. You you get the idea there. But here comes the kicker. In TestDaT, there are additional functions to organize your tests even further. And this is getting into a paradigm that admittedly is not quite comfortable to me just yet called behavior driven development or BDD syntax and test that for I'm not sure how many releases up to this point has spurred the use of a function called a set of functions called describe and it.
So in this last snippet of code, Jacob has an overall describe call that just simply says median. And then within it are a series of it calls and then the description in these it function calls is kind of the behavior itself that he's trying to test, where we have about the same value if it's only one element, the average of 2 values if it has an even number, the middle value in sequential order if it's an odd number of elements, and that it's the same value for ordered and unordered elements. I've seen this once before when I in Pasa Comps in recent years, I've had some interesting conversation with Martin Frigard, who has written a book called Shiny app as packages or something to that effect. But he has a specific chapter on behavior driven development in in light of specifications and requirements and how your test that test can kinda mirror that approach.
Admittedly, I have not had enough practice of describing it, no pun intended, in this workflow, but maybe it is quite helpful in your use case to have, again, another approach to how you organize related expectations into an overall testing block. So it is kind of a paradigm shift. Right? We could have either the nesting approach that we saw last week with the test that and multiple test that calls inside or the describe, and then these it functions are gonna have the expectations directly. Which is right for you? I don't know. Mike, what's your take on this?
[00:23:34] Mike Thomas:
This is the 2nd week in a row, I think, maybe 2 out of 3 weeks where I have found things in the test that package that I had no idea. Literally no idea existed. And, Eric, as I am, I know you are, we're very organized people and probably fairly opinionated about how we organize our code as well. I am open to this. I think if you were looking for a hot take on why we shouldn't consider this and and why the old way is the best, I'm not sure you're gonna get it from me right now. And it might be still because I'm I'm reeling from, learning about this new describe it paradigm.
But just like I was sort of blown away by the organization in the in the blog post around, nesting your tests last week or the week before. I'm similarly kinda blown away by this additional functionality here where we can leverage this describe it paradigm. I'm actually even more sort of not necessarily blown away, but but more keen on the the final sentence of this blog post. It says, if you wanna push test readability even further, check out how we can use the arrange, act, assert pattern to achieve that. And if you take a look at that link it's another nice short blog post as well by by Jacob. And, I I really like sort of the concept around how he he specifies arranging, the inputs sort of acting and and evaluating those inputs, and then passing those evaluated values to a set of test assertions after the fact. And, again, in that blog post, he's usually I know this this one isn't part of the the rweekly, but it's it's a cousin. It's close enough.
Again, he's leveraging that describe it paradigm. I think it's sort of I don't know. The more I look at it, it looks like it's it's sort of, you know, 6 in 1, half dozen in the other. I'm not sure how much of a difference it it makes. I I think it's just a matter of of preference, you know, if somebody on my team decided that they wanted to use describe it versus, you know, the traditional test that, verb from the test that package. I don't think I would I would care too much either way because I think it's it's legible both ways. I think it's fairly similar, in approach. It's it's sort of just different verbiage, if you will.
But it's very interesting for me, you know, to to know that this exists, and see that potentially maybe there there might be a use case for me in the future to try to adopt them. I'm just gonna stick one toe in the water, I think, for now. You know, I was taking a look at the some of the most recent packages that Hadley Wickham has put together, Elmer being one of them. And I wanted to take a look at the unit tests in there to see if Hadley was leveraging, you know, the older type of framework that traditional test that framework or or leveraging this describe and it paradigm.
And it looks like Hadley is still utilizing the old ways, if you will. So not necessarily adopted the describe it functionality, but I believe he has or had at one point in time a large hand in, authoring and making these decisions around the the test that package. Don't quote me on that. So I imagine perhaps, he was involved in maybe a pull request or 2 that that included this describe it functionality. So I was interested to see, you know, if the if he was going to be one of the folks who had also adopted sort of this new paradigm for code organization purposes purposes because I know that Hadley preaches, you know, good software development practices and and trying to articulate your code as as best as possible.
So I guess at the end of the day here, interested to see that we have these two new functions that I did not know about in the test that package. Am I all in on it? No. But I'm not all out on it either.
[00:27:38] Eric Nantz:
And I've I'm giving this the old college try as they say with this, app at work that I've just put in the production now. I took this behavior driven approach, describe its syntax for the the test. I will admit it it felt more cumbersome than right initially, but I'm thinking the payoff is not so much for me personally in the short term. The payoff hopefully is if I get additional developers to help me with this app as a, quote, unquote, scales, buzzword for all you industry folks out there, where I might need some help to orient somebody relatively quickly over. It's a dedicated member of my team or if it's contracting resource and whatnot.
And so being able to read that more verbose, but yet under you know, might say legible, might say more digestible way of organizing the test, then, hopefully, it makes it easier for them to write tests in that similar framework. And then we all can kinda have a common, you might say, style that we can gravitate towards for that particular project. Not too dissimilar to the whole paradigm of a given project having the same code style for all of its, you know, coding files, which, again, I can speak from experience when teams don't adhere to that. There'd be dragons when you do code reviews, and I won't say that for another time. That's my hot tech.
But I I think this is I think for future proofing things, I could see this being quite valuable, especially in the other context that I was dealing with. Right? I tried to I tried to have an attempt back to Martin's book that I'm gonna put in the show notes here of really articulating those user stories if you wanna use the agile methodology into what I'm actually trying to accomplish in that test. So if there's a, quote, unquote, project manager that wants to see how I how I assess all those different user stories even though the basically, that project manager is me for this particular project. But let's say, hypothetically, it was a more larger scale project. It was a good practice to see how it goes. So, again, it didn't feel comfortable yet, but maybe the proof is a year from now. So ask me a year from now, Mike. We'll see if I take changes.
[00:29:52] Mike Thomas:
Put it on my calendar. Oh, I don't think so.
[00:30:09] Eric Nantz:
And over the course of the last, you know, might say year or or so, we have seen an influx of really top notch resources and discussions in the in the r community as a whole with respect to the data dot table package. It's been around for many years. But thanks to recent NSF grant, they have had some real robust efforts to get the word out and also putting in the different situations and different context so that users that are new to the package can really understand what are the benefits of data dot table in their daily workflows. And so our last highlight is coming from the our data table community blog, which we've talked about in previous highlights.
In this case, the post is coming from Toby Dylan Hocking, who is a statistical researcher and the director of the LASSO lab at the University of Sherbrooke, which I believe is in Canada. And he has a very comprehensive post here about the comparison of data dot table functions for reshaping your data as compared to 2 other additional packages in your ecosystem. We're gaining a lot of momentum lately, which are DuckDV and Polars. So first, let's set the stage a little bit here because especially if you're very familiar with with database operations or if you're not so much like I was before, say, a few years ago, there is somewhat different terminology between what we use and, like, the tidyverse type of explanations of reshaping or in general data manipulation and the database equivalence of this. So when we talk about a long format, we are talking about not many columns but many rows in your dataset.
And often, there are groups of these rows based on, say, a variable type or a feature or whatnot. In SQL lingo, that's called unpivoted. That was new to me a while ago. Versus the wide format when you have one record, but then the columns are representing maybe different variables, and they are literally specific to that variable. You have, again, many columns, potentially fewer rows. That is called pivoted in the SQL lingo. Again, new to me, but that's where we're we're operating on where a lot of times, you would see in the community blog posts about benchmarks talking about the classical SQL like operations, like filtering, adding new variables, group summaries, or whatnot. But now we're literally turning it on its head, so to speak, by looking at the different ways we can change our data layout.
So the first part of the post is talking about just how data dot table accomplishes this, and this terminology is actually familiar from, I would say, previous language and dplyr for some of these other operations. But first, we will talk about going from a wide format to a long format and that unpivoting operation, if you will. And in data dot table, there's a function called melt for that, which if you're familiar with tidr back in the day, there was functions called melt and cast, I believe, which has now been changed to pivot longer and pivot wider, but that may be familiar for the tidr veterans out there. What was interesting about data dot table's take on this on this melt function is, in this example based on the iris dataset, when you deter when you want to say what are your variable that are variable or variables determining kind of the grouping of these observations, you can actually have multiple columns generated at once.
So in this example here, going from the the iris dataset, which has in wide format columns like sepal. Width, length sepal. Width, petal. Length, petal. Width, this melt function is taking a measure. Vars argument where you can feed in this measure function the names of 1 or more variables that are defined in these groupings and either a separator in the original name of the column or, down later in the example, regex to get to those column names. That, I must say, is quite handy and eliminates a post processing step, which, as Toby talks about later on, you need additional post processing to accomplish that that same step in both polars and DuckDV.
So there's already kind of a nice concise syntax, you might say, advantage at least at first glance with data dot table going from wide to long. And it's got some example visuals of what you might do with that data. But next, how do we accomplish this in Polars? And if you haven't heard of Polars before, this is a new binding to Rust for very efficient database data like operation and tidying operations. And yes, they do support reshaping or re pivoting these data sets as well. And that is a function called unpivot going back to that SQL language. But in the example snippet, you will see that at that the first attempt you cannot do more than one variable for that grouping of the observation.
So you would have to do that in post processing afterwards, which is not difficult. But, again, it's just an extra step. But, of course, it can be done. Lastly, for DuckDV via SQL queries, you can use the unpivot SQL keyword and then feeding into what are the variables that are going in, what are what's the name of the variable that's going to define that grouping of the long observations, what's the value the column name that you wanna use as, like, the numeric result. Once again, in this case, DuckDb cannot do that that nice convenience of 2 variables at once.
You have to do that in post processing as well. So there's example snippets and and Toby's code of the blog post here to talk about that additional post processing. That that is, again, all achievable. There is another example where you can reshape into multiple columns, and that might be helpful for different types of visualizations you want to conduct. And in this case, doing an additional column for or additional columns both for sepal and petal, but then there's a grouping variable called dim, which determines if it's the length or width. So data dot table has another way of doing that as well.
Again, the measure function comes into play there. You can see the example in the in the blog post. And then comes the comparisons for this operation. This is where my eyes open a little bit. So Toby does, a call to a an interesting func a package called a time. I'm not as familiar with this as I am with, like, the benchmark or a bench package, but he is taking a set of n values representing the number of rows in this, like, fictitious high dimensional Iris data set doing just some resampling and runs 3 different functions: 1 with the DuckDb on pivot, 2nd with the polars on unpivot, and lastly with data dot table and melt.
And across these cases, the ggplot that's put in the blog post clearly illustrates that the data dot table approach is definitely faster in this initial benchmark here than either of the other ones, especially as the end values get large. In particular, this was surprising to me, DuckDb had the worst performance by a pretty healthy margin when the end values got to, like, a 1000 rows and above. Now, again, maybe this isn't so surprising if DuckDb, of course, is based on the column type representation of data. Maybe it just isn't as optimized for these transposing operations. That very well could be a play. I'm still learning the ropes on the internals of it.
But that was interesting on top of the convenience function that data dot table has with MELT to get those multiple variables at once. It is showing, at least in this benchmark, a speedier process, especially as the number of rows increase. Now, again, maybe practically speaking, that won't be a huge impact to your workflows, but it was thought provoking nonetheless. But he does acknowledge that there's a lot of confounding in these comparisons, so it may not be quite apples to apples because of the different post processing that comes into play. And then when that is taking place, there are some additional visualizations when he kind of teases that out a bit that show the differences in even more, more fashion than what you saw in the previous plot. So interesting thought provoking exercise, but that's not the only operation, Mike, because we can also go the other way around. We can go from long to wide. So why don't you take us through that journey here? Exactly. If you are,
[00:39:50] Mike Thomas:
for some strange nonanalytics reason, looking to take your nice long data and reshape it into to wider data. And I shouldn't say that. There are some R packages that maybe it makes more sense to have each, you know, sort of sort of, a wider dataset, you know, representing your categorical data in multiple columns instead of a single column. But for me, this is a use case, that I face less often than going from from wide to long. But for those particular use cases where you do need to go long to wide, the function that you're gonna be using from data dot table is called dcast, and they provide some great examples here, Toby does, of how exactly to do that. Again, you can leverage a separator, if you would like to do that.
In this case, they're using that that period separator again, and the code is is fairly similar, to what you saw in the Melt code and allows you to pivot that that long column into a wider data frame, with multiple columns in it. If you are using polars, the method that you're going to be using there is literally called pivot. And, again, you know, the syntax is is pretty similar. You're sort of doing the opposite of what you did, when you were going from wide to long format. And then lastly, if you are using DuckDb, you're going to again use, the same named function as in Polaris. It's it's called pivot, the pivot command, which can be used to, as they say, recover the original iris data, the way that it is in wider format as opposed to long format.
And the SQL there is is pretty straightforward to look at especially compared to what we just walked through on the unpivot side of the equation. So drum roll, what everybody cares about here, right, is the benchmark comparison analysis. And this one goes a little bit inverse of the, wide to long approach such that, the engine, if you will, that has the best performance appears to be DuckDb, then closely followed by polars and then followed thereafter by data dot table. These benchmark lines that they have here are all pretty tight. I I wanna mention that that there's probably, you know, a much tighter a much tighter gaps between the 3 benchmarks here, across the 3 different engines, compared to what we saw in the wide to long approach.
So your experience is probably gonna be minimal, up until you start really scaling up to to datasets larger than, it looks like, you know, 24,000,000 rows roughly is is what DuckDV was able to handle in 0.1, seconds or or 10 milliseconds, if you will. So that was, you know, interesting to see sort of the benchmark there flip. One of the things that, Eric, I wanted to talk about here that I was curious about is I know and this is from listening to, especially, a recent podcast, not in the super data science podcast, with the author of the Polars Library, and I know DuckDV has some of this too, is both Polars and I think DuckDV have an option to do sort of a what's called a query plan and maybe lazy execution of, you know, chained functions that you might have together. Right? So we were talking about before where you can't do, you know, both in an unpivot and, you know, use a a separator in a value to split out additional columns all in a single function. You would have to chain that together in a couple different functions, in polars.
And I believe, you know, the way that polars works is depending on the default, and don't quote me on this, you know, it may default to what's called, like, eager evaluation, which is, you know, actually running those functions sort of in the order that they are written. But it provides you the option through some sort of binary flag to evaluate, your your query, if you will, or the code that you've written lazily, which means that behind the scenes, polars will put together a an optimal query plan that it believes will execute your code in the the most efficient and and fastest, if you will, way possible. And I imagine that DuckDb does some of this too.
And I also imagine that that query plan may take some time, right, under the hood to execute. So if we're only looking at these benchmarks up to 0.1 seconds, you know, I'd be interested in taking a look at datasets that are are maybe even larger, right, in the order of, like, a 100000000 rows, something like that or or larger. And seeing, you know, how these benchmarks compare after that at 0.1 second threshold to see if things change after the query planning, you know, algorithm is essentially run and the the time that it takes to do that has finished. And then we're really, you know, sort of just running, the actual code to or the actual query to do some of this data reshaping. So I'd be interested to see these benchmarks, I guess, you know, a little bit further out, extrapolate it a little bit further out in both time and magnitude of the dataset.
But, just an interesting thought that I had upon reading this blog post and it's it's super interesting to me, you know, where these, different engines sort of beat one another, if you will. You know, at the end of the day, if if you can't wait the extra 0.1 seconds or or or whatever it is, you know, half a second, just because you wanna use your your favorite engine. I guess it depends on your use case if you're standing up an API or something like that and you need response times as quick as possible and the data needs to be pivoted or or unpivoted, this is a great blog post to check out. But in sort of a general analytics, you know, approach here to to your day to day analysis.
The really cool thing I think about the ecosystem right now is is benchmarks are are getting to be fairly negligible, across most normal sized datasets.
[00:46:04] Eric Nantz:
That's a very, very fair assessment. And also with the advent of our computing technology with the processes processor cores going up and everything like that, we are seeing that for a data science workflow where let let's be real here. As much as the advancements in cloud happen, we know a lot of data science happens on people's laptops as well that with these compiled, you know, you know, I say back ends, wherever going with data dot table really based in c, and then you got polars based in Rust, the number, you know, getting a lot of attention in terms of performance. And, of course, DuckDb with its bindings. You are seeing this experience of these type of datasets. You you've got great choices here. And on that topic, like, query planning, if you want a way in especially in the DuckDV landscape, kind of get to what that query language actually looks like, I'll put a plug in the show notes for the Duckplier package, which had a CRAN release a few months ago because they have a handy function called explain that literally shows in kind of a plain text printout all the different operations that are about to be conducted. But it's doing it in an optimized order, I believe, to get the best performance as DuckDBC's fit for that particular operation, which then you can use with your familiar dplyr verbs of, like, mutate and summarize and whatnot. Again, those kind of classical operations that we've seen, you know, for for those type of work. So that was an interesting learning for me.
I do think the concept of the different data layouts is a frontier that I haven't seen as much attention on. So I'm certainly appreciative of Toby's post here to highlight, you know, the different ways that this can be accomplished. And I think, again, your use case may determine different things. I want, you know, for my back ends to have very minimal dependencies. And with that, to be able to fold into potentially a Shiny app for very quick operations about hogging the user's memory, for that particular app session. Those are my biggest, might say, criteria, if you will, as I'm looking at these.
And so I may have a use case where DuckDV does exactly what I need. There may be another use case for data dot table is king based on existing workflows. So, again, choice is gray here. Like you, Mike, I would love to see this expand in some of the different benchmarks and different scenarios to see where that plays out. But you can experiment with all these and kinda see what best fits for you. But, again, really comprehensive post by Toby here. And, again, it really shows that this funding that they're getting from this NSF grant is being put to great use to spread the word out about these different the different ways that we can accomplish these very common tasks in data science.
Well, we're we're running a bit well on time here on this episode. So we're gonna wrap things up after that great discussion of the highlights. But if you wanna get in touch with us further, we have multiple ways of doing that. You can send us a quick note in a contact page in our episode show notes. You can also send us a fun little boost along the way if you have a modern podcast app, or you can get in touch with us on social media. I am mostly on Mastodon these days, Yvette R podcast at podcastindex.social. Also find me on LinkedIn. Search my name and find me there. And, Mike, working on wisdom has got a hold of you.
[00:49:35] Mike Thomas:
Yep. You can find me on Mastodon at mike_thomas@fostodon.org, or you can find me on LinkedIn by searching Catchbrook Analytics, k e t c h b r o o k, to see what I'm up to lately.
[00:49:48] Eric Nantz:
Awesome stuff, my friend. Thanks again for a great episode, and thanks again for all of you tuning in from wherever you are. Have a great day, everybody, and we'll hopefully see you back here next week.